Registering Samples¶
Interactive¶
For this, we will use the materials in the samples/interactive/ folder of the example data. Here, we have the same spreadsheet in 2 different formats :
sample_spreadsheet.xlsx: this is in Excel format and has been annotated with colour-coding to highlight important featuressample_spreadsheet.tsv: tab-separated version of the above spreadsheet - this is the format accepted by Webin
In both cases, each row represents a sample and each column represents a metadata field.
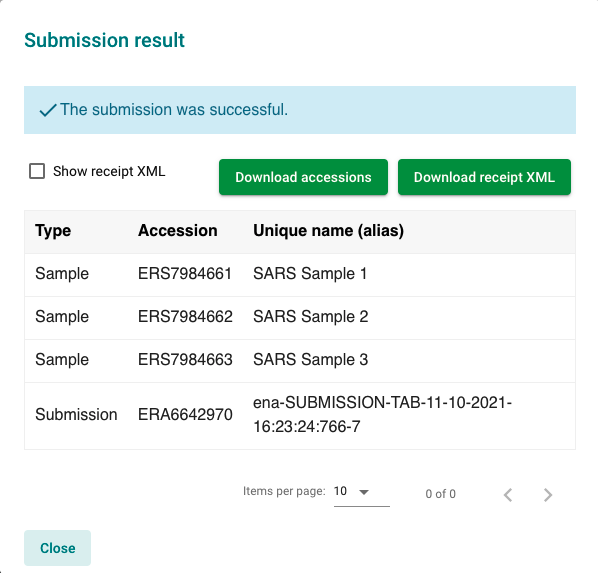
To upload, simply visit the Webin Submissions Portal again and click on the green ‘Register Samples’ button. Choose to ‘Upload filled spreadsheet’ and upload the example sheet provided: samples/interactive/sample_spreadsheet.tsv. You should get a popup showing a successful submission and your 3 new sample accessions.
Programmatic¶
In this section, we will use the materials in the samples/programmatic folder of the example data. Here, you will find several XML files. Navigate to the directory to see the file list:
cd $WORKSHOP
cd samples/programmatic
ls
samples.xml: this contains the same set of samples as those submitted interactively in the previous section.submission.xml: this XML is the same as that used to submit your study. It defines the<ADD/>action to create new samples.submission_modify.xml: this submission XML defines the<MODIFY/>action, to allow us to update existing samples.
Samples XML¶
The samples XML format allows us to define many samples inside a <SAMPLE_SET> tag. Each sample (enclosed in <SAMPLE> tags), contains:
<TITLE>tags : defining the title of the sample<SAMPLE_NAME>tags : defining the taxonomic information<DESCRIPTION>tags : providing a description of what’s been sampled andmany
<SAMPLE_ATTRIBUTE>tags : defining all other metadata fields
Note
Sample aliases are defined within the <SAMPLE> tag, e.g. <SAMPLE alias='this_alias'>.
In the example data, the alias has been suffixed with the word ‘programmatic’. This is to avoid clashes with the same samples
that were submitted interactively in the previous section.
Aliases must be unique.
Submit the samples¶
As we did with study registration, let’s send the samples XML and submission XML (with the <ADD/> action) to our test service
using cURL to perform a submission:
curl -u username:password -F "SUBMISSION=@submission.xml" -F "SAMPLE=@samples.xml" "https://wwwdev.ebi.ac.uk/ena/submit/drop-box/submit/"
Again, you should receive a receipt XML with information about submission success and accession numbers. Note that this time, you will receive a <SAMPLE> tag for each submitted sample (in this case, 3). Please take note of each sample alias and accession as we will use these later to submit data files against.
For more general information on programmatic sample registration, please see our documentation.
Modifying a sample¶
Sometimes, erroneous metadata can be uploaded, and the sample needs to be updated at a later date. This can be achieved by editing the sample XML file to update the relevant fields, and resubmitting with a submission XML containing the <MODIFY/> action in place of <ADD/>.
First, open the
samples.xmlfile and update a metadata field of your choice. e.g. new collection date. Save the file.This time, we will submit with the
submission_modify.xml, which instructs the service to update an existing sample. The update uses the alias to detect existing samples, so it is important not to change the alias.
curl -u username:password -F "SUBMISSION=@submission_modify.xml" -F "SAMPLE=@samples.xml" "https://wwwdev.ebi.ac.uk/ena/submit/drop-box/submit/"
Check the receipt for successful update. Note, it will also report the samples that haven’t been updated.
Warning
Although sample metadata can be updated, these updates are not automatically propagated to the EMBL files of their sequences. This is due to computational constraints.
Updating samples will be reflected in the sample page in the ENA browser, the BioSamples record, and the COVID-19 Data Portal.
If it is very important for your EMBL files to be updated with new metadata, please contact our helpdesk at virus-dataflow@ebi.ac.uk and we will endeavour to assist you.
Tip
Now we have our samples registered to our project, it’s time to add some data files. We’ll start with submitting raw read data.